A Benefit / Feature
Service Goals
With the rapid development of biotechnology, protein drugs have become an important means of treating various diseases. However, the development process of protein drugs faces many challenges, such as long screening time and low success rate. In order to solve these problems, we focus on providing high-throughput de novo protein drug design services and protein drug screening software services.
De novo protein drug design services:Using artificial intelligence technology, we can carry out efficient de novo protein drug design based on the structural and functional characteristics of specific disease targets. This service is designed to help customers quickly find protein drug candidates with high activity and low toxicity, thereby accelerating the drug development process.
Protein drug screening software services: In view of the shortcomings of existing protein drug screening methods, we developed an efficient and accurate protein drug screening software. The software can accurately predict the activity and performance of drug candidates through rapid simulation and evaluation of a large number of protein molecules and disease target structures. Customers can use the software platform to conduct large-scale, high-throughput drug screening, which greatly improves screening efficiency and accuracy.
Service
De novo Protein Design
Protein design is a fundamental aspect of protein engineering with extensive applications, like antibodies, peptides, and enzymes. Protein design has three steps: specify hotspot binding sites, protein backbone generation, and fix-backbone sequence design
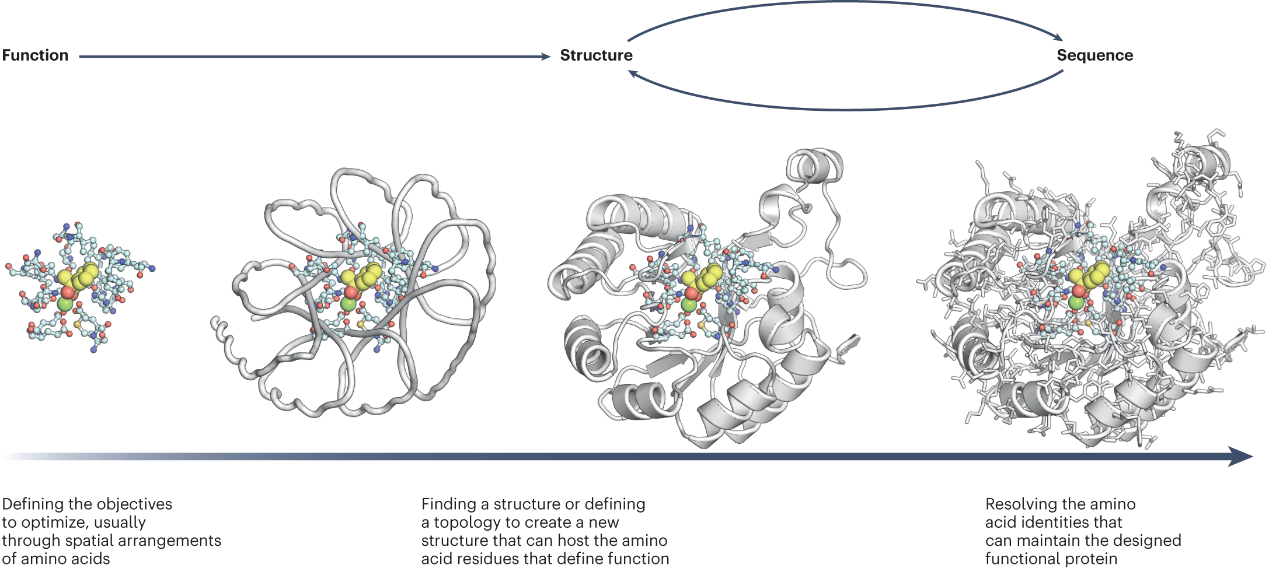
Figure 1. The workflow of protein design: specify a desired function, protein backbone generation, and fix-backbone sequence design(1).
Hotspot binding sites search/prediction
Identifying these hotspots is essential for understanding protein function and designing protein. IMO Medical Technology Company used ScanNet for detecting protein–protein and protein–antibody binding sites from 3D structure.
Protein Backbone Generation
Protein Backbone Generation Methods
Protein backbone generation can be set into two different types: unconditional generation and conditional generation (motif-scaffolding). Intelligent Medicine Original (IMO) Medical Technology Company has used the protein language model as supervision to develop the GPDL model for precise and efficient motif-scaffolding design. GPDL exhibited the most superior success rate among the 24 universally used cases for the motif-scaffolding task.

Figure 2. The framework of the GPDL(7)
Critical Assessment of Protein Backbone Generation Methods
IMO Medical Technology Company has developed a unified framework for the assessment of backbone generation methods. This framework system evaluated unconditional generation and motif-scaffolding. The representative protein backbone generation methods based on their performance and availability are shown in Table 1.
Unconditional Generation
IMO Medical Technology Company evaluated unconditional generation across metrics like designability, diversity, novelty, efficiency and structural properties. FrameFlow and Genie performs the best on short proteins, while RFdiffusion shows an outstanding performance upon generating long proteins.
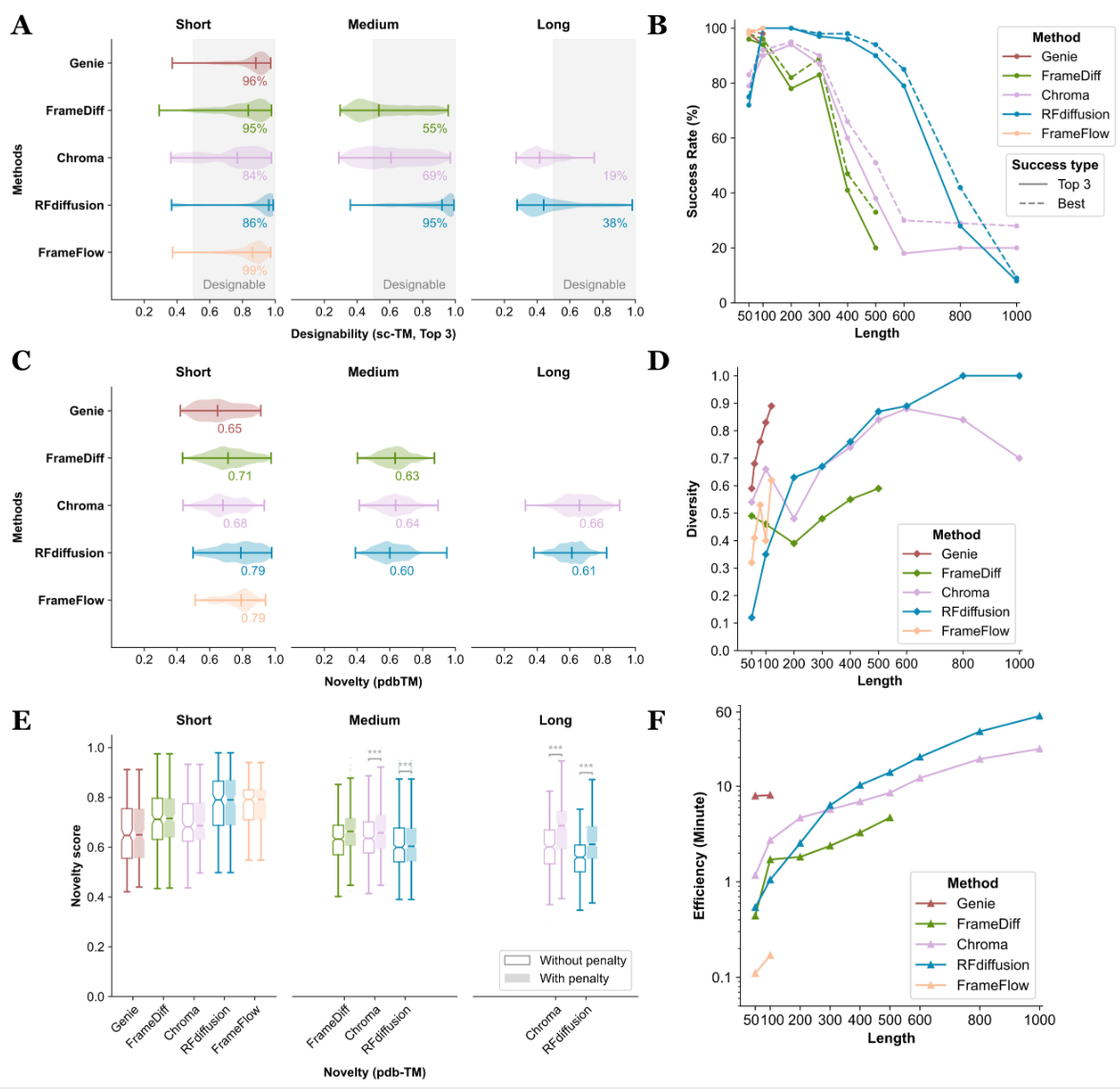
Figure 3. Evaluation results of unconditional generation.
Motif-scaffolding
For conditional generation, IMO Medical Technology Company conducted a comprehensive experiment across the 24 cases universally used for motif-scaffolding task. GPDL exhibited the most outstanding performance with an average success rate .
Fixed Backbone Generation Protein Sequence Design
Fixed Backbone Generation Protein Sequence Design Methods
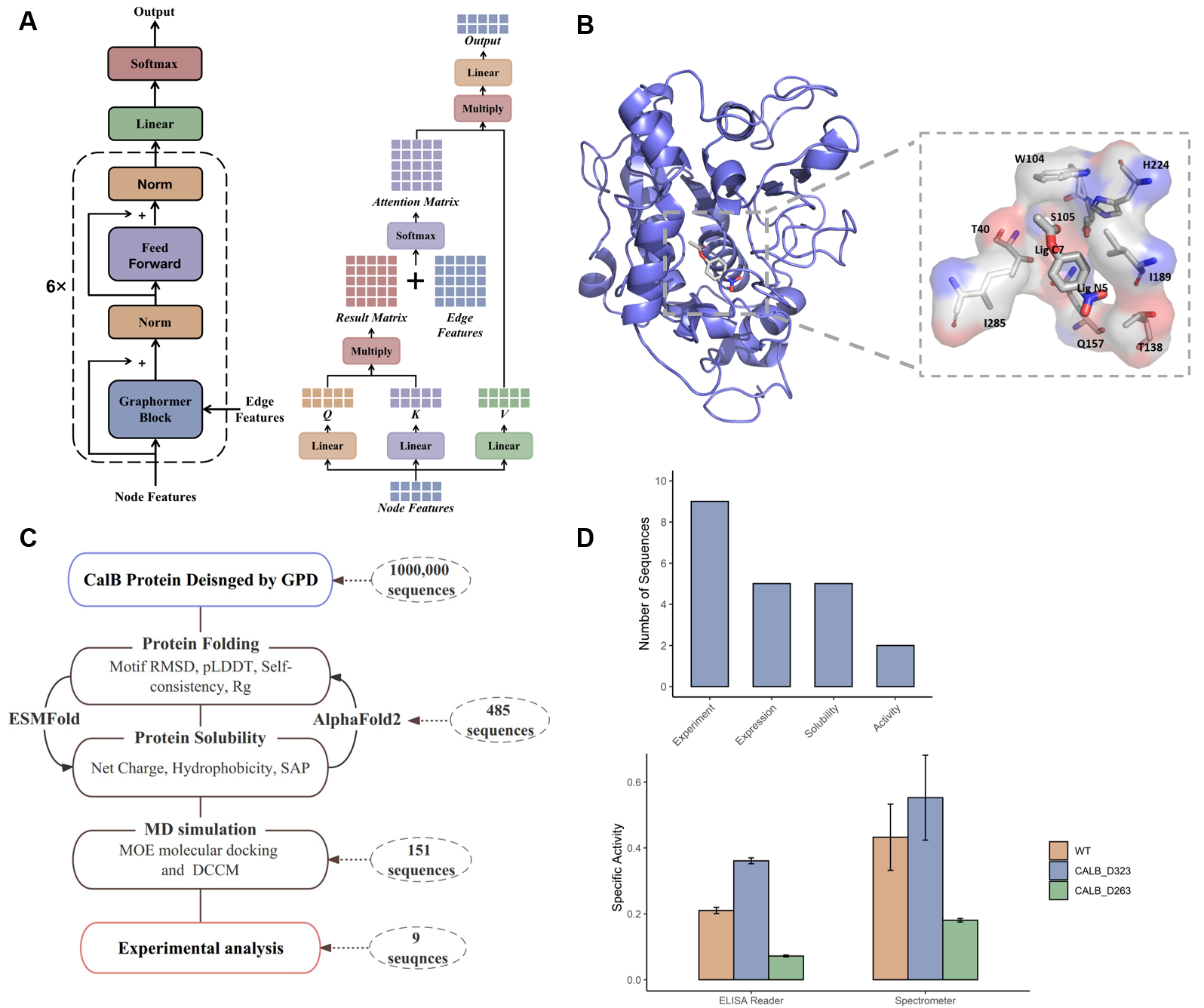
Figure 5. the GPD architecture and the design workflow of CalB hydrolase. (A) The overall architecture of GPD. (B) The CalB and substrate (p-nitrophenyl acetate C2) complex after MD simulation and the region of CalB active sites is enlarged. (C) The design workflow of CalB including CalB sequences design, protein folding ability, protein solubility and MD simulation. (D) The specific activity of designed sequences and wild type using enzyme linked immunosorbent assay (ELISA) Reader and Spectrometer.
Critical Assessment of Protein Sequence Design Methods
IMO Medical Technology Company have developed a multi-indicator comparative evaluation framework for sequence design methods using a diverse set of indicators that cover several important aspects, including sequence recovery, diversity, root-mean-square deviation of protein structure, secondary structure, and the distribution of polar and non-polar amino acids. As a result, ProteinMPNN consistently maintained its leading position, while Structured Transformer outperformed GPD and PiFold.
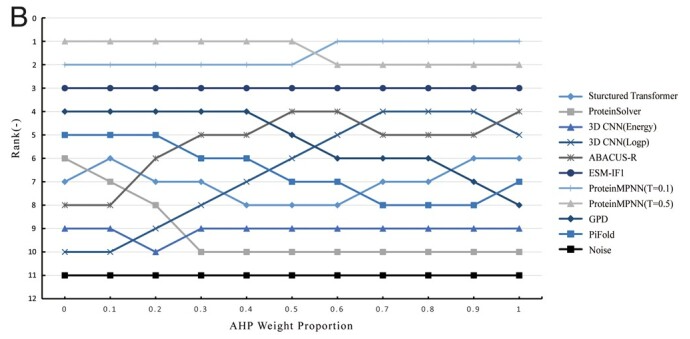
Figure 6. The ranking results. The ranking of 11 objects changes with the weight proportion of AHP.
Graft Design
Dual agonists design such as GLP/GLP-1 design.
Protein Binder Design
Protein binders design refers to the process of designing proteins given a target and specification of interface hotspot residues. IMO Medical Technology Company used RFdiffusion for binder design.
Peptide Design
Design Peptide Based on Fixed Backbone Design
Peptide design plays a pivotal role in therapeutics. The common steps for peptide design are: (1) AI for peptides generation; (2) Functional screening; (3) wet laboratory validation(13-14). IMO Medical Technology Company have redesigned the GLP-1. The first round Pull-down experiment tests demonstrate that 42/60 artificially-designed proteins has binding affinity with GLP-1R. Remarkably, 74% (31/42) of active designed GLP-1 exhibited superior binding affinity compared to Simaglutide.
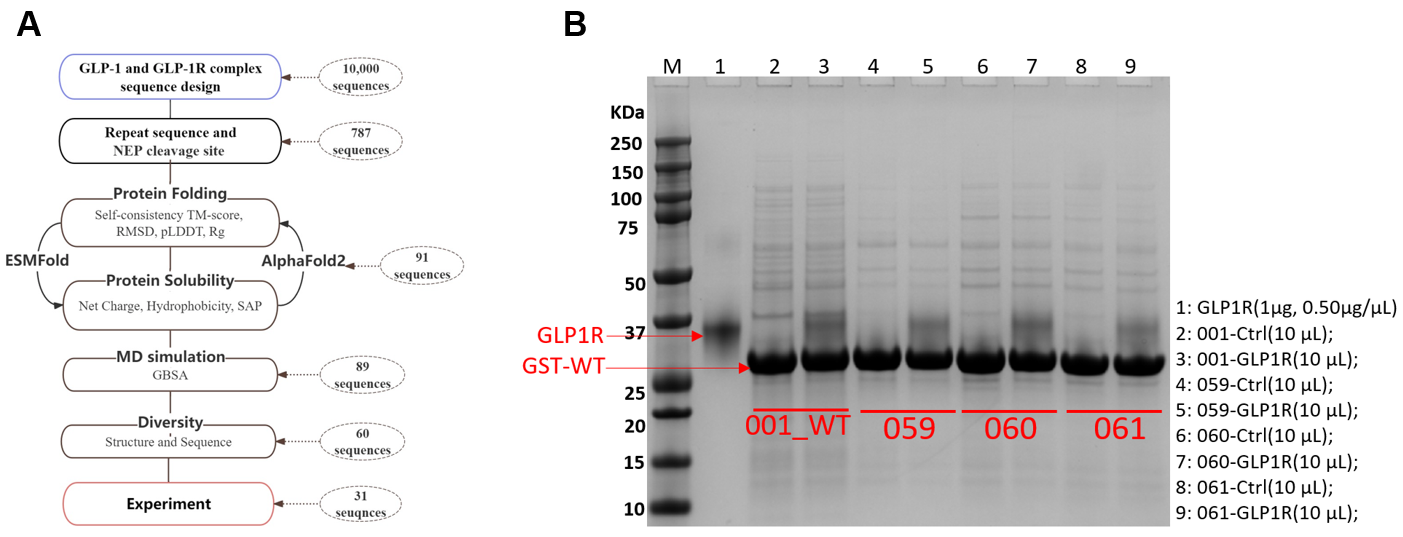
Figure 7. The design workflow of GLP-1. (A) The design workflow of GLP-1 including GLP-1 sequences design, protein folding ability, protein solubility, MD simulation, and sequence diversity. (B) The first round Pull-down experiment of designed GLP-1 compared with Simaglutide (wildtype, WT).
Peptide Sequence-Structure Co-design
For peptides design, IMO Medical Technology Company used sequence-structure co-design models for designing peptide binders for given protein targets with three dimensional and binding site information.
Non-natural Amino Acids and Cyclization
Non-natural amino acids include the substitution of L-amino acids by D-amino acids and natural amino acid analogues. MD simulations were carried out for the non-natural amino acids peptide structure. ff03CMAP force field was used for simulation.
Loop Design
Loop design is difficult because there no efficient methods to predict accurately the interaction strengths between the designed peptides and target exhibiting considerable conformational flexibility (intrinsically disordered region). For example, the designing peptide inhibitors for NEMO. Using IDP force field, we could accurately predict the binding affinity.
Enzyme-substrate Design
Enzyme Design Based on Fixed Backbone Design
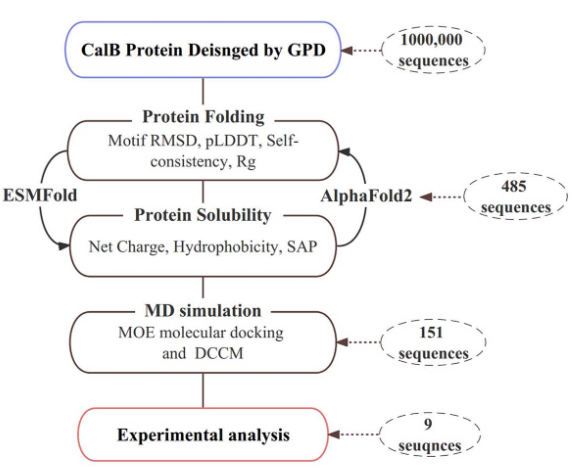
The Intelligent Medicine Original (IMO) Medical Technology Company has developed GPD model for fixed backbone sequence design. GPD was employed to design Candida antarctica lipase B (CalB) enzyme. Nine sequences that fulfilled the catalytic mechanism were chosen for specific activity analysis using an ELISA Reader. The experimental results show a 1.7-fold increase in catalytic activity compared to that of the wild-type CalB and strong substrate selectivity on p-nitrophenyl acetate with different carbon chain lengths.
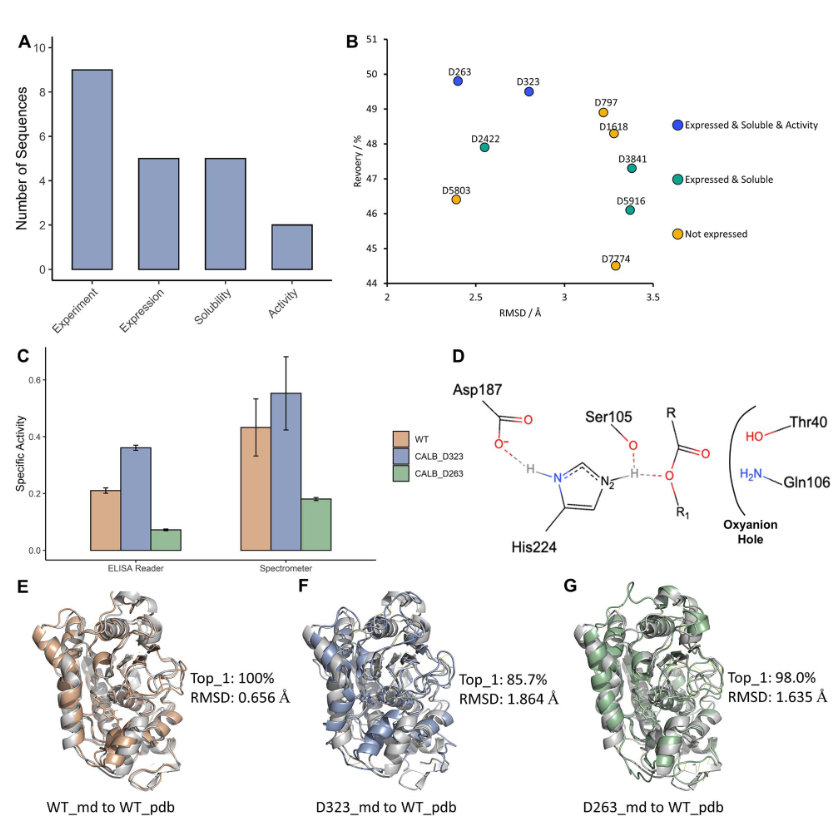
Figure 9. CalB experiments results of designed sequences. (A) The number of designed sequences for experimented and expressed. (B)The RMSD and recovery values of designed sequences. (C) The specific activity of designed sequences and wild type using enzyme linked immunosorbent assay (ELISA) Reader. (D) The model of pre-catalytic state of CALB-substrate complex. (E) The alignment between MD simulation clustered wild type (WT) structure and WT pdb structure. (F) The alignment between MD simulation clustered D263 structure and WT pdb structure.
Antibody-Antigen Design
IMO Medical Technology Company has developed a unified framework for the assessment of antibody design methods, including backbone and all-atom methods.
VHH Nanobody
A VHH nanodrug refers to a therapeutic agent that is based on VHH antibodies, also known as nanobodies. These are single-domain antibodies derived from the unique heavy-chain-only antibodies found in camelids (like llamas and alpacas) and certain species of sharks. VHH nanodrugs have potential applications in various fields, including molecular imaging, diagnostics, drug delivery, and cancer immunotherapy.
Computational Filtering
Folding ability, Solubility, and Functionality
IMO Medical Technology Company have developed a workflow for functional screening of designed sequences. The designed sequences were virtually screened on the basis of protein folding ability, protein solubility and molecular dynamics (MD) simulation.
Druggable
Stability
IMO Medical Technology Company are developing a deep learning model to unravel those peptide properties with most impact on proteolytic stability and thus potential t1/2 predicting ability(24). IMO Medical Technology Company also used ADMETlab3.0 to predict half-life of peptides(25). ADMETlab3.0 websereve is widely used to evaluate the absorption, distribution, metabolism, excretion, and toxicity of molecular.
Membrane Permeability
The membrane permeability of peptide drugs depends on multiple factors, including peptide length and amino acid composition. Membrane permeability is an essential indicator of oral bioavailability and intracellular targeting. IMO Medical Technology Company are developing a deep learning-based membrane permeability prediction model with reliable generalization performance.
Immunogenicity/Anti-Drug Antibodies
The immunogenicity of peptides is a critical consideration in drug development, as it can lead to reduced drug efficacy, altered pharmacokinetics, and adverse immune reactions. Therefore, assessing and mitigating the immunogenicity risk of therapeutic peptides is an essential part of the drug development process. IMO Medical Technology Company used DeepImmuno-CNN and diffRBM to predict immunogenicity.
