Recently, Professor Haifeng Chen, co-founder of company, developed an innovative and efficient new protein sequence design method called GPD. Compared to the current state-of-the-art method, proteinMPNN, GPD exhibits significantly higher sequence diversity and generates sequences 2.2 times faster, greatly enhancing the de novo design capabilities of industrial enzymes and protein drugs. The research results were published in the CAS top journal “Briefings in Bioinformatics.”

Protein design is central to almost all protein engineering problems because it enables the creation of proteins with novel biological functions and can improve the catalytic efficiency of enzymes, among other benefits. A key issue in protein design is the fixed-backbone protein sequence design, which aims to design new sequences that conform to a predetermined protein backbone structure. However, existing sequence design methods have various limitations, such as low sequence diversity and insufficient experimental validation of the designed functional proteins, which severely hinder functional protein design.
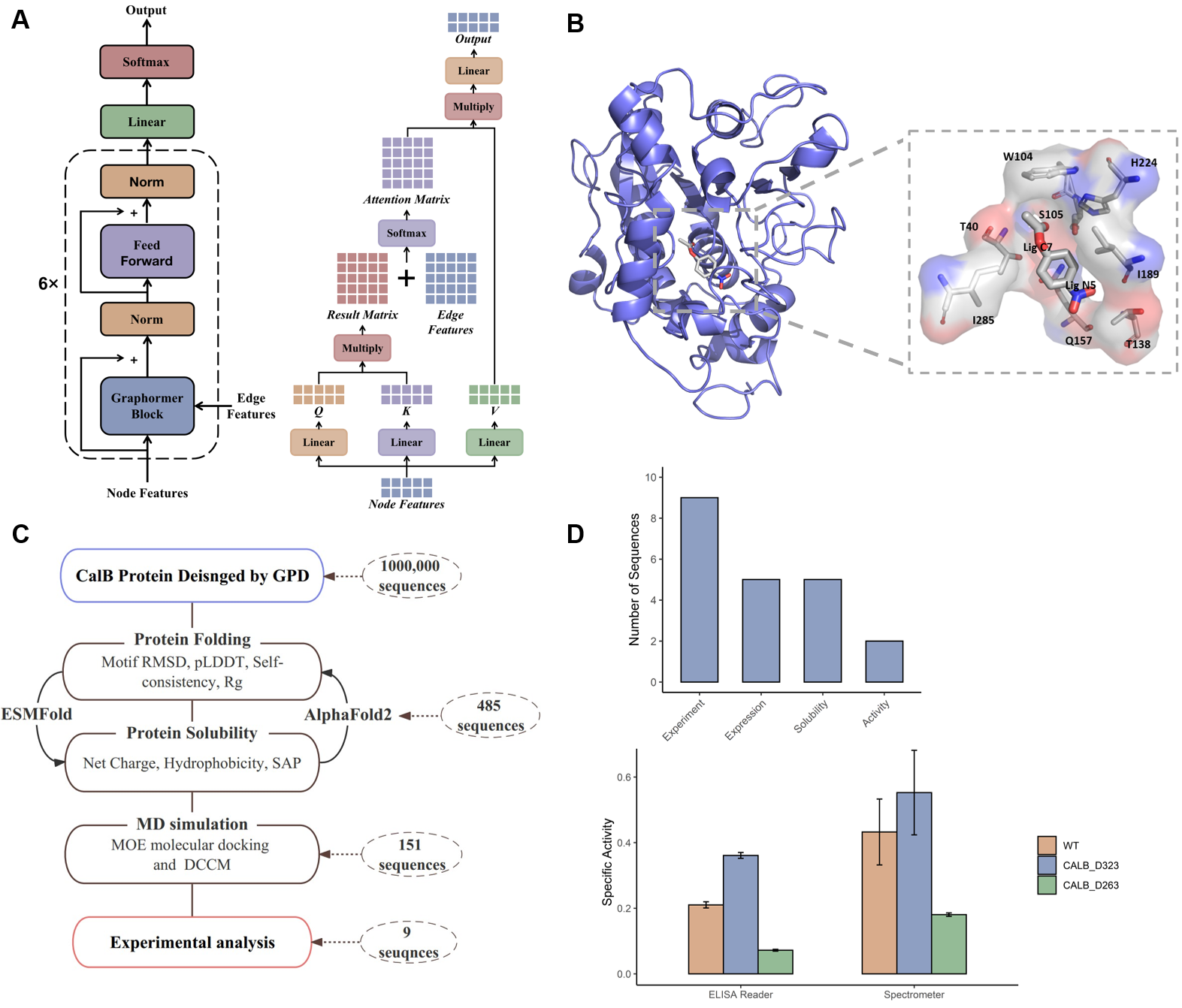
To address these limitations, the team developed the Graphormer-based Protein Design (GPD) model in this study. This model uses a Transformer to perform graph-based 3D protein structure representation, incorporating Gaussian noise and sequence random masking into node features, thereby enhancing the quality of sequence design.
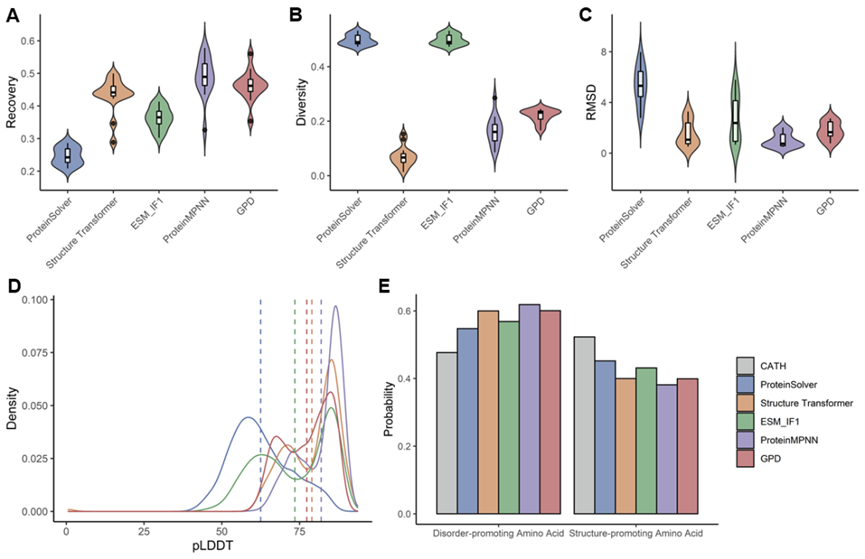
Subsequently, the team evaluated the sequence design quality of GPD during the research process and found that it could design and generate more reasonable protein sequences while maintaining high sequence diversity. Most of the designed sequences could also fold into the desired structures in structural prediction models. Overall, GPD outperformed existing models in terms of sequence foldability, sequence homology, and sequence diversity.
Additionally, Intelligent Medicine Original, in collaboration with Shanghai Jiao Tong University, applied GPD to the redesign of Antarctic yeast lipase (CALB), generating and screening nine artificially designed protein sequences. Compared to the wild-type CalB, one of the designed sequences exhibited a 1.7-fold increase in catalytic activity. The experimental results further demonstrate the rationality of GPD’s design, as well as its efficiency over previous rational design or directed evolution methods.
Moreover, enzyme activity tests for multiple substrates revealed that the sequences designed by GPD exhibited high substrate specificity, showing strong substrate selectivity on p-nitrophenyl acetate with different carbon chain lengths (C2-C16). This has significant implications for the industrial application of CALB enzymes.
Intelligent Medicine Original’s innovative protein sequence design method, GPD, can be used for the de novo design of industrial enzymes and protein drugs, providing a methodological foundation for the rapid development of new productive forces. The company aims to introduce advanced computational methods into the biopharmaceutical field, striving to create an AI-powered platform for protease modification and innovative drug design.
